-Search query
-Search result
Showing 1 - 50 of 196 items for (author: li & yh)

EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-35192: 
A cryoEM structure of the dimer of (S)-carbonyl reductase II
Method: single particle / : Li YH, Wang C, Zhang RZ, Xu Y, Hunt JF
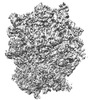
EMDB-34866: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
Method: single particle / : Wang YH, Zhou J

EMDB-42481: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42482: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D

EMDB-42483: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-2heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42485: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42486: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D

PDB-8ur5: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8ur7: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-40787: 
Negative stain EM map 1 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40788: 
Negative stain EM map 2 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40792: 
Negative stain EM map 3 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40793: 
Negative stain EM map 4 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40794: 
Negative stain EM map 5 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40795: 
Negative stain EM map 6 of polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40798: 
Negative stain EM map 1 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40800: 
Negative stain EM map 2 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40801: 
Negative stain EM map 3 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40802: 
Negative stain EM map 4 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40831: 
Negative stain EM map 5 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40832: 
Negative stain EM map 6 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40833: 
Negative stain EM map 1 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40834: 
Negative stain EM map 2 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40835: 
Negative stain EM map 3 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40836: 
Negative stain EM map 4 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40837: 
Negative stain EM map 5 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40838: 
Negative stain EM map 6 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40839: 
Negative stain EM map 7 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40840: 
Negative stain EM map of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40841: 
Negative stain EM map 2 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40842: 
Negative stain EM map 3 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40843: 
Negative stain EM map 4 of polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40844: 
Negative stain EM map 1 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-40845: 
Negative stain EM map 2 of polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon
Method: single particle / : Dosey A, King NP

EMDB-35775: 
The rice Na+/H+ antiporter SOS1 in an auto-inhibited state
Method: single particle / : Zhang XY, Tang LH, Zhang CR, Nie JW

EMDB-35950: 
The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state
Method: single particle / : Zhang XY, Tang LH, Zhang CR, Nie JW
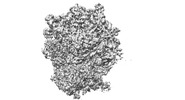
EMDB-34867: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
Method: single particle / : Wang YH, Zhou J

EMDB-34868: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
Method: single particle / : Wang YH, Zhou J

EMDB-34864: 
Cryo-EM Structures and Translocation Mechanism of Crenarchaeota Ribosome
Method: single particle / : Wang YH, Zhou J

EMDB-34372: 
CryoEM structure of the RAD51_ADP filament
Method: helical / : Miki Y, Luo SC, Ho MC

EMDB-36040: 
Composite map of the Rad51-ADP filament
Method: helical / : Miki Y, Luo SC, Ho MC

EMDB-34710: 
Cryo-EM structure of WeiTsing
Method: single particle / : Qin L, Tang LH, Chen YH

EMDB-16147: 
Cryo-EM structure of the human SIN3B histone deacetylase complex at 3.7 Angstrom
Method: single particle / : Wan MSM, Muhammad R, Koliopolous MG, Alfieri C

EMDB-16148: 
Cryo-EM structure of the human SIN3B histone deacetylase core complex at 2.8 Angstrom
Method: single particle / : Wan MSM, Muhammad R, Koliopolous MG, Alfieri C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
